The meiofauna as neglected carriers of antibiotic resistant and pathogenic bacteria in freshwater ecosystems
Accepted: 22 November 2021
HTML: 87
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Authors
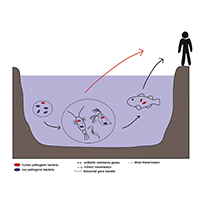
The World Health Organization considers antibiotic resistance as one of the main threats to human and other animals' health. Despite the measures used to limit the spread of antibiotic resistance, the efforts made are not enough to tackle this problem. Thus, it has become important to understand how bacteria acquire and transmit antibiotic resistant genes (ARGs), in particular in the environment, given the close connection between the latter and human and animal health, as defined by the One-Health concept. Aquatic ecosystems are often strongly impacted by anthropogenic activities, making them a source for ARGs and antibiotic resistant bacteria (ARB). Although freshwater meiofauna have been the object of active research, few studies have focused on the relationship between the spread of antibiotic resistance and these organisms. In this review, we investigated freshwater meiofauna as carriers of resistances since they play a central role in the aquatic environments and can harbor human and animal potential pathogens. We assessed if these animals could contribute to the spread of ARGs and of potentially pathogenic bacteria. Only four taxa (Rotifera, Chironomidae, Cladocera, Copepoda) were found to be the subject of studies focused on antibiotic resistance. The studies we analyzed, although with some limitations, demonstrated that ARGs and ARB can be found in these animals, and several of them showed the presence of potentially pathogenic bacteria for humans and animals within their microbiome. Thus, meiofauna can be considered a source and a reservoir, even if neglected, of ARGs and ARB for the freshwater environments. However, further studies are needed to evaluate the impact of the meiofauna on the spread and persistence of antibiotic resistance in these ecosystems.
Supporting Agencies
Cariplo Foundation (WARFARE project, grant n° 2018-0995), International Commission for the Protection of Italian-Swiss Waters (“INDAGINI LIMNOLOGICHE SUL LAGO MAGGIORE” program)How to Cite

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Similar Articles
- Reinaldo L. BOZELLI, Maurizio TONSI, Fabrizio SANDRINI, Marina MANCA, A Big Bang or small bangs? Effects of biotic environment on hatching , Journal of Limnology: Vol. 67 No. 2 (2008)
- Uwe SELIG, Manfred MICHALIK, Thomas HÜBENER, Assessing P status and trophic level of two lakes by speciation of particulate phosphorus forms , Journal of Limnology: Vol. 65 No. 1 (2006)
- Giuseppe MORABITO, Delio RUGGIU, Pierisa PANZANI, Recent dynamics (1995-1999) of the phytoplankton assemblages in Lago Maggiore as a basic tool for defining association patterns in the Italian deep lakes , Journal of Limnology: Vol. 61 No. 1 (2002)
- Alcide CALDERONI, Gabriele A. TARTARI, Evolution of the water chemistry of Lake Orta after liming , Journal of Limnology: Vol. 60 No. 1 (2001)
You may also start an advanced similarity search for this article.
-
Andrea Di Cesare, Raffaella Sabatino, Tomasa Sbaffi, Diego Fontaneto, Diego Brambilla, Andrea Beghi, Franca Pandolfi, Cristina Borlandelli, Davide Fortino, Giovanni Biccai, Pietro Genoni, Gianluca CornoChemosphere : 2023

 https://doi.org/10.4081/jlimnol.2021.2054
https://doi.org/10.4081/jlimnol.2021.2054





