Mercury methylation in oxic aquatic macro-environments: a review
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Authors
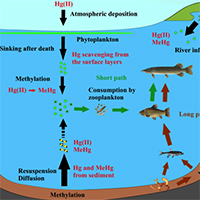
Mercury methylation in aquatic environments is a key process that incorporates this neurotoxin into the food chain and ultimately the human diet. Mercury methylation is considered to be essentially biotic and mainly driven by sulfate-reducing bacteria present in the bottom sediments in aquatic systems. However, in recent decades, many researchers have shown that this methylation also occurs in oxic layers in conjunction with a high content of particulate organic matter and localized depletion of dissolved oxygen. The goals of this review are to summarize our current understanding of Hg methylation in water columns of both marine and freshwater environments, as well as to highlight knowledge gaps and future research needs. Most of the literature showed that suspended particles (known as marine and lake snow) could be the microenvironment in which Hg methylation could occur across oxic water columns, because they have been recognized as a site of organic matter mineralization and as presenting oxygen gradients around and inside them. To date, the majority of these studies concern marine environments, highlighting the need for more studies in freshwater environments, particularly lacustrine systems. Investigating this new methylmercury production environment is essential for a better understanding of methylmercury incorporation into the trophic chain. In this review, we also propose a model which attempts to highlight the relative importance of a MeHg epilimnetic path over a MeHg benthic-hypolimnetic path, especially in deep lakes. We believe that this model could help to better focus future scientific efforts in limnic environments regarding the MeHg cycle.
Edited by
Diego Fontaneto, CNR-IRSA, Verbania, ItalyHow to Cite

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






